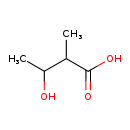
Showing metabocard for 2-Methyl-3-hydroxybutyric acid (BMDB0000354)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2016-09-30 22:29:10 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2020-04-22 15:03:07 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BMDB ID | BMDB0000354 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | 2-Methyl-3-hydroxybutyric acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | 2-Methyl-3-hydroxybutyric acid, also known as 3-hydroxy-2-methylbutyric acid or 2-methyl-3-hydroxybutanoate, belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. 2-Methyl-3-hydroxybutyric acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. 2-Methyl-3-hydroxybutyric acid is a potentially toxic compound. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C5H10O3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight | 118.1311 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight | 118.062994186 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 3-hydroxy-2-methylbutanoic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | nilic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 473-86-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CC(O)C(C)C(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C5H10O3/c1-3(4(2)6)5(7)8/h3-4,6H,1-2H3,(H,7,8) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | VEXDRERIMPLZLU-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty acids and conjugates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Hydroxy fatty acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ontology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected but not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofunction | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Application | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biospecimen Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0000354 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer Compound ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB021977 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | 141015 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Compound ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | 3786 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound | 160471 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 37051 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Inoue, Yoshio; Sano, Fumitaka; Nakamura, Kazuhiro; Yoshie, Naoko; Saito, Yuji; Satoh, Hiroyasu; Mino, Takashi; Matsuo, Tomonori; Doi, Yoshiharu. Microstructure of copoly(3-hydroxyalkanoates) produced in the anaerobic-aerobic activated sludge process. Polymer International (1996), 39(3), 183-9. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||