| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:33:05 UTC |
|---|
| Update Date | 2020-05-11 20:08:53 UTC |
|---|
| BMDB ID | BMDB0000577 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 5beta-Coprostanol |
|---|
| Description | 5beta-Coprostanol, also known as coprosterol or 5β coprostanol, belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. Thus, 5beta-coprostanol is considered to be a sterol. Based on a literature review a significant number of articles have been published on 5beta-Coprostanol. |
|---|
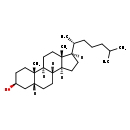
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3beta,5beta)-Cholestan-3-ol | ChEBI | | 5beta Coprostanol | ChEBI | | 5beta-Cholestan-3beta-ol | ChEBI | | Coprosterol | ChEBI | | (3b,5b)-Cholestan-3-ol | Generator | | (3Β,5β)-cholestan-3-ol | Generator | | 5b Coprostanol | Generator | | 5Β coprostanol | Generator | | 5b-Cholestan-3b-ol | Generator | | 5Β-cholestan-3β-ol | Generator | | 5b-Coprostanol | Generator | | 5Β-coprostanol | Generator | | 3b-Hydroxy-5b-cholestanol | HMDB | | Stercorin | HMDB | | 5 beta-Cholestan-3 beta-ol | HMDB | | 5 alpha Cholestan 3 alpha ol | HMDB | | Coprostanol | HMDB | | 5 beta-Cholestan-3 alpha-ol | HMDB | | Cholestan 3 ol | HMDB | | Cholestanol, (3alpha, 5beta)-isomer | HMDB | | beta-Cholestanol | HMDB | | beta-Cholestan-3 beta-ol, 5 | HMDB | | Cholestan-3-ol | HMDB | | Cholestanol | HMDB | | Dihydrocholesterol | HMDB | | beta-Ol, 5 beta-cholestan-3 | HMDB | | 5 alpha Cholestan 3 beta ol | HMDB | | beta Cholestanol | HMDB | | 5 alpha-Cholestan-3 alpha-ol | HMDB | | 5 alpha-Cholestan-3 beta-ol | HMDB | | 5 beta Cholestan 3 beta ol | HMDB |
|
|---|
| Chemical Formula | C27H48O |
|---|
| Average Molecular Weight | 388.6694 |
|---|
| Monoisotopic Molecular Weight | 388.370516158 |
|---|
| IUPAC Name | (1S,2S,5S,7R,10R,11S,14R,15R)-2,15-dimethyl-14-[(2R)-6-methylheptan-2-yl]tetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-5-ol |
|---|
| Traditional Name | (1S,2S,5S,7R,10R,11S,14R,15R)-2,15-dimethyl-14-[(2R)-6-methylheptan-2-yl]tetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-5-ol |
|---|
| CAS Registry Number | 360-68-9 |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])[C@]3([H])CC[C@]4([H])C[C@@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCCC(C)C |
|---|
| InChI Identifier | InChI=1S/C27H48O/c1-18(2)7-6-8-19(3)23-11-12-24-22-10-9-20-17-21(28)13-15-26(20,4)25(22)14-16-27(23,24)5/h18-25,28H,6-17H2,1-5H3/t19-,20-,21+,22+,23-,24+,25+,26+,27-/m1/s1 |
|---|
| InChI Key | QYIXCDOBOSTCEI-NWKZBHTNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Cholestane steroids |
|---|
| Direct Parent | Cholesterols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cholesterol-skeleton
- Cholesterol
- 3-beta-hydroxysteroid
- Hydroxysteroid
- 3-hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Membrane
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 102 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-052u-9527000000-d23f4273340e8c85dc8a | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-052u-9527000000-d23f4273340e8c85dc8a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05i0-0109000000-761dcf3fe040f44fa80e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0002-3104900000-654dcbe9dd07b5a589f2 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - EI-B (JEOL JMS-01-SG-2) , Positive | splash10-052u-9527000000-d23f4273340e8c85dc8a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0079-0009000000-57023400d5e478a8c746 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dr-3039000000-9ce6c0a362b9f376d3f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ab9-4159000000-32959ebb75235db63ac3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-134a888410f8beddca3b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0009000000-c42f43f5e21141f5a909 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ab9-1009000000-8096d56c7bfdd221f9c2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-34ef429285e58bb4b314 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0009000000-780ac33d63315f658159 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-0009000000-2b8e0e9916832dc47876 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0009000000-44afea742dee80d0734e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9032000000-166905331dad000292e6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4j-9820000000-f066461902c4fe39e2d4 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|