| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:35:00 UTC |
|---|
| Update Date | 2020-05-11 20:55:08 UTC |
|---|
| BMDB ID | BMDB0000693 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Heparan sulfate |
|---|
| Description | Heparan sulfate, also known as heparitin or alpha-idosane, belongs to the class of organic compounds known as disaccharide sulfates. These are disaccharides carrying one or more sulfate group on a sugar unit. Based on a literature review a significant number of articles have been published on Heparan sulfate. |
|---|
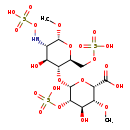
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Heparan sulfuric acid | Generator | | Heparan sulphate | Generator | | Heparan sulphuric acid | Generator | | alpha-Idosane | HMDB | | Heparan N-sulfate | HMDB | | Heparan N-sulphate | HMDB | | Heparatan sulfate | HMDB | | Heparatan sulphate | HMDB | | Heparitin | HMDB | | Heparitin monosulfate | HMDB | | Heparitin monosulphate | HMDB | | Heparitin sulfate | HMDB | | Heparitin sulphate | HMDB | | HHS 5 | HMDB | | N-Acetylheparan sulfate | HMDB | | N-Acetylheparan sulphate | HMDB | | Suleparoid | HMDB | | Tavidan | HMDB | | (2S,3R,4R,5S,6R)-4-Hydroxy-6-{[(2S,3R,4S,5S,6R)-4-hydroxy-6-methoxy-5-[(sulfooxy)amino]-2-[(sulfooxy)methyl]oxan-3-yl]oxy}-3-methoxy-5-(sulfooxy)oxane-2-carboxylate | HMDB | | (2S,3R,4R,5S,6R)-4-Hydroxy-6-{[(2S,3R,4S,5S,6R)-4-hydroxy-6-methoxy-5-[(sulphooxy)amino]-2-[(sulphooxy)methyl]oxan-3-yl]oxy}-3-methoxy-5-(sulphooxy)oxane-2-carboxylate | HMDB | | (2S,3R,4R,5S,6R)-4-Hydroxy-6-{[(2S,3R,4S,5S,6R)-4-hydroxy-6-methoxy-5-[(sulphooxy)amino]-2-[(sulphooxy)methyl]oxan-3-yl]oxy}-3-methoxy-5-(sulphooxy)oxane-2-carboxylic acid | HMDB |
|
|---|
| Chemical Formula | C14H25NO21S3 |
|---|
| Average Molecular Weight | 639.52 |
|---|
| Monoisotopic Molecular Weight | 639.008120351 |
|---|
| IUPAC Name | (2S,3R,4R,5S,6R)-4-hydroxy-6-{[(2S,3R,4S,5S,6R)-4-hydroxy-6-methoxy-5-[(sulfooxy)amino]-2-[(sulfooxy)methyl]oxan-3-yl]oxy}-3-methoxy-5-(sulfooxy)oxane-2-carboxylic acid |
|---|
| Traditional Name | (2S,3R,4R,5S,6R)-4-hydroxy-6-{[(2S,3R,4S,5S,6R)-4-hydroxy-6-methoxy-5-[(sulfooxy)amino]-2-[(sulfooxy)methyl]oxan-3-yl]oxy}-3-methoxy-5-(sulfooxy)oxane-2-carboxylic acid |
|---|
| CAS Registry Number | 9050-30-0 |
|---|
| SMILES | CO[C@@H]1O[C@@H](COS(O)(=O)=O)[C@H](O[C@@H]2O[C@@H]([C@H](OC)[C@@H](O)[C@@H]2OS(O)(=O)=O)C(O)=O)[C@@H](O)[C@@H]1NOS(O)(=O)=O |
|---|
| InChI Identifier | InChI=1S/C14H25NO21S3/c1-29-9-7(17)10(35-38(23,24)25)14(34-11(9)12(18)19)33-8-4(3-31-37(20,21)22)32-13(30-2)5(6(8)16)15-36-39(26,27)28/h4-11,13-17H,3H2,1-2H3,(H,18,19)(H,20,21,22)(H,23,24,25)(H,26,27,28)/t4-,5-,6-,7+,8-,9+,10-,11-,13+,14+/m0/s1 |
|---|
| InChI Key | AUQASCCBXZRMEG-RHKLHVFKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as disaccharide sulfates. These are disaccharides carrying one or more sulfate group on a sugar unit. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Disaccharide sulfates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Disaccharide sulfate
- 1-o-glucuronide
- O-glucuronide
- Glucuronic acid or derivatives
- Glycosyl compound
- O-glycosyl compound
- Sulfuric acid ester
- Alkyl sulfate
- Sulfate-ester
- Sulfuric acid monoester
- Pyran
- Oxane
- Organic sulfuric acid or derivatives
- Secondary alcohol
- Acetal
- Carboxylic acid derivative
- Carboxylic acid
- Dialkyl ether
- Ether
- Monocarboxylic acid or derivatives
- Oxacycle
- Organoheterocyclic compound
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Alcohol
- Organonitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_17) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_18) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0kai-9400000000-404fa85a4342eafffaec | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9400000000-eec3b32ccab8e708c47b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001i-9400000000-ad221bda41e892ec4944 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dl-0029057000-552f350b2eda79515ae3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fk9-1079141000-0b28aff1bbd9804b9bea | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05fr-1394000000-5fa03687957fac5d0d21 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00kr-2923734000-0b76c8fcf07de6f623f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-4903331000-2468d6710f7d484c4bc2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00xr-7901000000-aa930e030c48c4c24016 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000019000-61194d75acd7b76eeb63 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000b-9380064000-507abf4005bb44c55d09 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-9232401000-8b2d59e928ea5f7d2bf8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000009000-37a3f7caf057b5f5103f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-3061394000-8c7732b97f391e56ce66 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pb9-3291100000-baca1b5774273b60adc0 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, CD3OD, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|