| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:35:48 UTC |
|---|
| Update Date | 2020-04-22 15:05:05 UTC |
|---|
| BMDB ID | BMDB0000739 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Isodesmosine |
|---|
| Description | Isodesmosine, also known as Ide, belongs to the class of organic compounds known as tetracarboxylic acids and derivatives. These are carboxylic acids containing exactly four carboxyl groups. Based on a literature review a significant number of articles have been published on Isodesmosine. |
|---|
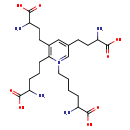
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-(4-Amino-4-carboxybutyl)-1-(5-amino-5-carboxypentyl)-3,5-bis(3-amino-3-carboxypropyl)pyridinium | ChEBI | | 2-Amino-6-[3,5-bis(3-amino-4-hydroxy-4-oxobutyl)-2-(4-amino-5-hydroxy-5-oxopentyl)pyridin-1-ium-1-yl]hexanoic acid | ChEBI | | Ide | ChEBI | | 2-Amino-6-[3,5-bis(3-amino-4-hydroxy-4-oxobutyl)-2-(4-amino-5-hydroxy-5-oxopentyl)pyridin-1-ium-1-yl]hexanoate | Generator | | 2-(4-Amino-4-carboxybutyl)-1-(5-amino-5-carboxypentyl)-3,5-bis(3-amino-3-carboxypropyl)-pyridinium | HMDB | | Iso-desmosin | HMDB |
|
|---|
| Chemical Formula | C24H40N5O8 |
|---|
| Average Molecular Weight | 526.6031 |
|---|
| Monoisotopic Molecular Weight | 526.287688281 |
|---|
| IUPAC Name | 3,5-bis(3-amino-3-carboxypropyl)-2-(4-amino-4-carboxybutyl)-1-(5-amino-5-carboxypentyl)pyridin-1-ium |
|---|
| Traditional Name | isodesmosine |
|---|
| CAS Registry Number | 991-01-5 |
|---|
| SMILES | NC(CCCC[N+]1=CC(CCC(N)C(O)=O)=CC(CCC(N)C(O)=O)=C1CCCC(N)C(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C24H39N5O8/c25-16(21(30)31)4-1-2-11-29-13-14(7-9-18(27)23(34)35)12-15(8-10-19(28)24(36)37)20(29)6-3-5-17(26)22(32)33/h12-13,16-19H,1-11,25-28H2,(H3-,30,31,32,33,34,35,36,37)/p+1 |
|---|
| InChI Key | RGXCTRIQQODGIZ-UHFFFAOYSA-O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as tetracarboxylic acids and derivatives. These are carboxylic acids containing exactly four carboxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Tetracarboxylic acids and derivatives |
|---|
| Direct Parent | Tetracarboxylic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Tetracarboxylic acid or derivatives
- Alpha-amino acid
- Alpha-amino acid or derivatives
- Aralkylamine
- Pyridine
- Pyridinium
- Heteroaromatic compound
- Amino acid or derivatives
- Amino acid
- Azacycle
- Organoheterocyclic compound
- Carboxylic acid
- Organic nitrogen compound
- Organic oxygen compound
- Organic oxide
- Organopnictogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Hydrocarbon derivative
- Amine
- Organic cation
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0019-0000900000-78ef29d9446bcb2f1511 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00di-1000059000-f1c087858d1fc630f361 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0000190000-808de3e96eaf83168632 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0095-0000910000-97e503d6c21442da41c7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-2300900000-99327764dbc9812bcc34 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000090000-62b53001eb9cde723507 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-1000090000-7d10eeb30c8bc5581672 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00dl-9200540000-2bad06401982c266a8c7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a5l-0000930000-e39fe63f78706dd677f3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-3102910000-74e46e64c33bcdc78925 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0540-7009700000-9b1b4fd5ade54caf2d8c | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|