| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:39:43 UTC |
|---|
| Update Date | 2020-05-21 16:29:27 UTC |
|---|
| BMDB ID | BMDB0001000 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | dUDP |
|---|
| Description | dUDP belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleoside diphosphates. These are pyrimidine nucleotides with a diphosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. dUDP is an extremely weak basic (essentially neutral) compound (based on its pKa). dUDP exists in all eukaryotes, ranging from yeast to humans. |
|---|
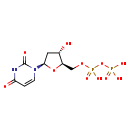
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2'-Deoxyuridine 5'-diphosphate | ChEBI | | 2'-Deoxyuridine 5'-diphosphoric acid | Generator | | 2'-Deoxyuridine-5'-diphosphate | HMDB | | Deoxyuridine-diphosphate | HMDB | | 2'-Deoxyuridine 5'-(trihydrogen diphosphate) | HMDB | | 2’-Deoxyuridine 5’-(trihydrogen diphosphate) | HMDB | | 2’-Deoxyuridine 5’-diphosphate | HMDB | | Deoxyuridine 5'-diphosphate | HMDB | | Deoxyuridine 5’-diphosphate | HMDB | | Deoxyuridine diphosphate | HMDB | | dUDP | HMDB |
|
|---|
| Chemical Formula | C9H14N2O11P2 |
|---|
| Average Molecular Weight | 388.1618 |
|---|
| Monoisotopic Molecular Weight | 388.007282324 |
|---|
| IUPAC Name | [({[(2R,3S,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3-hydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]phosphonic acid |
|---|
| Traditional Name | dUDP |
|---|
| CAS Registry Number | 4208-67-7 |
|---|
| SMILES | O[C@H]1C[C@@H](O[C@@H]1COP(O)(=O)OP(O)(O)=O)N1C=CC(=O)NC1=O |
|---|
| InChI Identifier | InChI=1S/C9H14N2O11P2/c12-5-3-8(11-2-1-7(13)10-9(11)14)21-6(5)4-20-24(18,19)22-23(15,16)17/h1-2,5-6,8,12H,3-4H2,(H,18,19)(H,10,13,14)(H2,15,16,17)/t5-,6+,8+/m0/s1 |
|---|
| InChI Key | QHWZTVCCBMIIKE-SHYZEUOFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleoside diphosphates. These are pyrimidine nucleotides with a diphosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine deoxyribonucleotides |
|---|
| Direct Parent | Pyrimidine 2'-deoxyribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine 2'-deoxyribonucleoside diphosphate
- Organic pyrophosphate
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyrimidine
- Alkyl phosphate
- Heteroaromatic compound
- Vinylogous amide
- Tetrahydrofuran
- Lactam
- Urea
- Secondary alcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cytoplasm
- Mitochondria
- Nucleus
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004m-6911000000-1f7795debc1eaf0802e6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-002g-9741200000-2a5ff084260317ace541 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-1900000000-3d46a12380fc537ed482 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-4910000000-e4d9c69da2a4f1af3f7d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-6900000000-38fc58820b65e6b77481 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-6309000000-e32a735e5de73376968b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056u-9801000000-189cbc835aaa1c3340b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9100000000-07b08a59c26df7bb2d1e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-aa2d362ee8b723b7d488 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9111000000-b30fec320d4d01519c03 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-5632c2fa3d2d7c954d90 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-3913000000-cbd6b90181ad2dcbef10 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-6910000000-514b52784d71478993f8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-4900000000-32241af48bd1f1fd6e37 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|