| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:43:26 UTC |
|---|
| Update Date | 2020-05-11 20:22:15 UTC |
|---|
| BMDB ID | BMDB0001272 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Nicotine glucuronide |
|---|
| Description | Nicotine glucuronide belongs to the class of organic compounds known as purine ribonucleoside triphosphates. These are purine ribobucleotides with a triphosphate group linked to the ribose moiety. Nicotine glucuronide is a very strong basic compound (based on its pKa). |
|---|
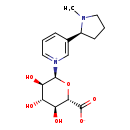
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-1-alpha-D-Glucopyranuronosyl-3-(1-methyl-2-pyrrolidinyl)pyridinium inner salt | ChEBI | | 3,4,5-Trihydroxy-6-[5-(1-methylpyrrolidin-2-yl)pyridin-1-yl]-oxane-2-carboxylate | ChEBI | | Nicotine N-glucuronide | ChEBI | | (S)-1-a-D-Glucopyranuronosyl-3-(1-methyl-2-pyrrolidinyl)pyridinium inner salt | Generator | | (S)-1-Α-D-glucopyranuronosyl-3-(1-methyl-2-pyrrolidinyl)pyridinium inner salt | Generator | | 3,4,5-Trihydroxy-6-[5-(1-methylpyrrolidin-2-yl)pyridin-1-yl]-oxane-2-carboxylic acid | Generator | | (S)-1-alpha-D-Glucopyranuronosyl-3-(1-methyl-2-pyrrolidinyl)-pyridinium inner salt | HMDB | | (S)-1-alpha-delta-Glucopyranuronosyl-3-(1-methyl-2-pyrrolidinyl)-pyridinium inner salt | HMDB | | (S)-1-alpha-delta-Glucopyranuronosyl-3-(1-methyl-2-pyrrolidinyl)pyridinium inner salt | HMDB | | 1-alpha-D-Glucopyranuronosyl-3-(1-methyl-2-pyrrolidinyl)-(S)-pyridinium | HMDB | | 1-alpha-D-Glucopyranuronosyl-3-[(2S)-1-methyl-2-pyrrolidinyl]-pyridinium | HMDB | | 1-alpha-delta-Glucopyranuronosyl-3-(1-methyl-2-pyrrolidinyl)-(S)-pyridinium | HMDB | | 1-alpha-delta-Glucopyranuronosyl-3-[(2S)-1-methyl-2-pyrrolidinyl]-pyridinium | HMDB | | Nicotine-glucuronide | HMDB |
|
|---|
| Chemical Formula | C16H22N2O6 |
|---|
| Average Molecular Weight | 338.3557 |
|---|
| Monoisotopic Molecular Weight | 338.147786446 |
|---|
| IUPAC Name | 1-[(2S,3R,4S,5S,6S)-6-carboxylato-3,4,5-trihydroxyoxan-2-yl]-3-[(2S)-1-methylpyrrolidin-2-yl]-1λ⁵-pyridin-1-ylium |
|---|
| Traditional Name | nicotine glucuronide |
|---|
| CAS Registry Number | 152306-59-7 |
|---|
| SMILES | CN1CCC[C@H]1C1=C[N+](=CC=C1)[C@H]1O[C@@H]([C@@H](O)[C@H](O)[C@H]1O)C([O-])=O |
|---|
| InChI Identifier | InChI=1S/C16H22N2O6/c1-17-6-3-5-10(17)9-4-2-7-18(8-9)15-13(21)11(19)12(20)14(24-15)16(22)23/h2,4,7-8,10-15,19-21H,3,5-6H2,1H3/t10-,11-,12-,13+,14-,15-/m0/s1 |
|---|
| InChI Key | SAWAIULJDYFLPD-SOAFEQHCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine ribonucleoside triphosphates. These are purine ribobucleotides with a triphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | Purine ribonucleoside triphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine ribonucleoside triphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Hydroxypyrimidine
- Alkyl phosphate
- Pyrimidine
- Monosaccharide
- Phosphoric acid ester
- N-substituted imidazole
- Organic phosphoric acid derivative
- Heteroaromatic compound
- Azole
- Imidazole
- Tetrahydrofuran
- Secondary alcohol
- 1,2-diol
- Oxacycle
- Organoheterocyclic compound
- Azacycle
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Alcohol
- Organic oxygen compound
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ab9-9363000000-498032dac1d25a048f1c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-000l-6695530000-5f3cb4632382491d1884 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0009000000-c0a6517967854466764f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004r-3209000000-9c96a94c455824fbcb27 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03xu-6911000000-358133c8eb23ea49be3b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-470460c450e2ce5dc00f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-5594000000-c8f1d538ca13bdab946d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9200000000-83503a4041514150d674 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0209000000-45aaad3735689e97aa1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03yr-0905000000-ca062e765a6a892950c2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-3910000000-d6d53f13a1744fd96f8a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0229000000-8425edbf57e8743a529b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06ri-5977000000-71e200566908d48e305d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fr-8941000000-0a955908878509d2a872 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|