| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:51:26 UTC |
|---|
| Update Date | 2020-05-11 20:23:08 UTC |
|---|
| BMDB ID | BMDB0002123 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 1,3,7-Trimethyluric acid |
|---|
| Description | 1,3,7-Trimethyluric acid, also known as 1,3,7-trimethyluric acid or 1,3,7-trimethyluric acid, belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. 1,3,7-Trimethyluric acid is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). 1,3,7-Trimethyluric acid exists in all living organisms, ranging from bacteria to humans. 1,3,7-Trimethyluric acid can be biosynthesized from caffeine; which is catalyzed by the enzymes cytochrome P450 1A2, cytochrome P450 3A4, cytochrome P450 2C8, cytochrome P450 2C9, and cytochrome P450 2E1. In cattle, 1,3,7-trimethyluric acid is involved in the metabolic pathway called the caffeine metabolism pathway. |
|---|
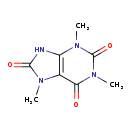
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,3,7-Trimethyl-2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione | ChEBI | | 1,3,7-Trimethylurate | ChEBI | | 7,9-Dihydro-1,3,7-trimethyl-1H-purine-2,6,8(3H)-trione | ChEBI | | 8-Oxocaffeine | ChEBI | | 8-Oxy-caffeine | ChEBI | | 1,3,7-Trimethylate | Generator | | 1,3,7-Trimethylic acid | Generator | | 1,3, 7-Trimethyluric acid | HMDB | | 1,3,7-Trimethyl-7,9-dihydro-1H-purine-2,6,8(3H)-trione | HMDB | | 2,6,8-Trihydroxy-1,3,7-trimethylpurine | HMDB | | 8-Hydroxy-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione | HMDB | | Trimethyl uric acid | HMDB |
|
|---|
| Chemical Formula | C8H10N4O3 |
|---|
| Average Molecular Weight | 210.19 |
|---|
| Monoisotopic Molecular Weight | 210.075290206 |
|---|
| IUPAC Name | 1,3,7-trimethyl-2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione |
|---|
| Traditional Name | 1,3,7-trimethyluric acid |
|---|
| CAS Registry Number | 5415-44-1 |
|---|
| SMILES | CN1C(=O)NC2=C1C(=O)N(C)C(=O)N2C |
|---|
| InChI Identifier | InChI=1S/C8H10N4O3/c1-10-4-5(9-7(10)14)11(2)8(15)12(3)6(4)13/h1-3H3,(H,9,14) |
|---|
| InChI Key | BYXCFUMGEBZDDI-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- N-substituted imidazole
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Urea
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 5.5 mg/mL at 15 °C | Not Available | | LogP | -0.37 | GASPARI,F & BONATI,M (1987) |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0udi-1900000000-7ceff209b1b0d4f92cf0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, N/A (Annotated) | splash10-03di-0090000000-c6489ab09ec74614db4c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, N/A (Annotated) | splash10-001a-6900000000-ac2e74a460c865c0636b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, N/A (Annotated) | splash10-067l-9100000000-cbac42dd288a8ef44fd8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0006-0900000000-a9bff7d94cd5ffe280b9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-0udj-0900000000-5316637dfdcce19f087a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0imj-3930000000-0593f0aaec496efc7696 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-000f-9500000000-fb28fd8903d55811b0a1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-000f-0910000000-eca560d36d14fb72dfe9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0190000000-6d4b9f5e8e42f3040e5e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-000l-7900000000-2ba0d4f876fceab5182f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-000f-9700000000-a42f3584a2bad7add22d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-052o-1920000000-53b778bbd3181f2ed83a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-000l-3900000000-4305cd286c077e521079 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0690000000-62dc7eb029289b61c3b1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-000f-0900000000-3eb9fdcfd2e67d0f4e2c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9100000000-f438ad2b70b2e9735643 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0190000000-42add8be62bbafed200c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03dj-2930000000-2855ca36942d5e8f069e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-066u-9200000000-708fd95bbe618c2ace7d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0090000000-153ea9ea8b4fe3692bfb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ik9-1790000000-2d17afbf34659471f524 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0m2a-4900000000-c783d40d7ed0c6ae44a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0190000000-9858aab04a40f6286f93 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-1790000000-2089332ab35983ee2c77 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-066u-8900000000-53f14da0e8f43679f03f | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|