| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:52:37 UTC |
|---|
| Update Date | 2020-05-11 20:04:11 UTC |
|---|
| BMDB ID | BMDB0002206 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Molybdopterin |
|---|
| Description | Molybdopterin belongs to the class of organic compounds known as pyranopterins and derivatives. These are pterin derivatives in which a pyran ring is fused either to the pyrimidine ring or the pyrazine ring of the pterin moiety. Molybdopterin is a moderately basic compound (based on its pKa). Molybdopterin exists in all living organisms, ranging from bacteria to humans. |
|---|
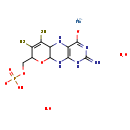
| Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C10H16MoN5O8PS2 |
|---|
| Average Molecular Weight | 525.31 |
|---|
| Monoisotopic Molecular Weight | 526.923197 |
|---|
| IUPAC Name | molybdenum(2+) ion 8-[(hydrogen phosphonooxy)methyl]-2-imino-6,7-disulfanyl-1H,2H,5H,5aH,8H,9aH,10H-pyrano[3,2-g]pteridin-4-olate dihydrate |
|---|
| Traditional Name | molybdenum(2+) ion 8-[(hydrogen phosphonooxy)methyl]-2-imino-6,7-disulfanyl-1H,5H,5aH,8H,9aH,10H-pyrano[3,2-g]pteridin-4-olate dihydrate |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | O.O.[Mo++].[H]C1(COP(O)([O-])=O)OC2([H])NC3=C(NC2([H])C(S)=C1S)C([O-])=NC(=N)N3 |
|---|
| InChI Identifier | InChI=1S/C10H14N5O6PS2.Mo.2H2O/c11-10-14-7-4(8(16)15-10)12-3-6(24)5(23)2(21-9(3)13-7)1-20-22(17,18)19;;;/h2-3,9,12,23-24H,1H2,(H2,17,18,19)(H4,11,13,14,15,16);;2*1H2/q;+2;;/p-2 |
|---|
| InChI Key | VUKICSJFFDCESC-UHFFFAOYSA-L |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyranopterins and derivatives. These are pterin derivatives in which a pyran ring is fused either to the pyrimidine ring or the pyrazine ring of the pterin moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Pterins and derivatives |
|---|
| Direct Parent | Pyranopterins and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyranopterin
- Secondary aliphatic/aromatic amine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyran
- Pyrimidine
- Alkyl phosphate
- Heteroaromatic compound
- Alkylthiol
- Oxacycle
- Secondary amine
- Thioenol
- Azacycle
- Organic transition metal salt
- Amine
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organosulfur compound
- Organic nitrogen compound
- Organonitrogen compound
- Organic zwitterion
- Organic salt
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0001490000-bf4dfec34731bf10ebad | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-1606490000-e83ef8869348bf49af28 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-020r-4982200000-b29a94b7f07e6ea22651 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-05e9-9606040000-926bcbb9296f4a9cd5e0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9101010000-2dd652c6c798de5919fb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-58324c83b689024dc935 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|