| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:01:25 UTC |
|---|
| Update Date | 2020-04-22 15:11:19 UTC |
|---|
| BMDB ID | BMDB0002721 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
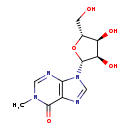
| Common Name | 1-Methylinosine |
|---|
| Description | 1-Methylinosine, also known as m(1)i, belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. Based on a literature review a significant number of articles have been published on 1-Methylinosine. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| m(1)I | ChEBI | | N1-Methylinosine | ChEBI | | 1-Methyl-inosine | HMDB |
|
|---|
| Chemical Formula | C11H14N4O5 |
|---|
| Average Molecular Weight | 282.2527 |
|---|
| Monoisotopic Molecular Weight | 282.096419578 |
|---|
| IUPAC Name | 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methyl-6,9-dihydro-1H-purin-6-one |
|---|
| Traditional Name | 1-methylinosine |
|---|
| CAS Registry Number | 2140-73-0 |
|---|
| SMILES | CN1C=NC2=C(N=CN2[C@@H]2O[C@H](CO)[C@@H](O)[C@H]2O)C1=O |
|---|
| InChI Identifier | InChI=1S/C11H14N4O5/c1-14-3-13-9-6(10(14)19)12-4-15(9)11-8(18)7(17)5(2-16)20-11/h3-5,7-8,11,16-18H,2H2,1H3/t5-,7-,8-,11-/m1/s1 |
|---|
| InChI Key | WJNGQIYEQLPJMN-IOSLPCCCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleosides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Purine nucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleoside
- Glycosyl compound
- N-glycosyl compound
- 6-oxopurine
- Hypoxanthine
- Pentose monosaccharide
- Purinone
- Imidazopyrimidine
- Purine
- Pyrimidone
- Pyrimidine
- N-substituted imidazole
- Monosaccharide
- Vinylogous amide
- Tetrahydrofuran
- Heteroaromatic compound
- Azole
- Imidazole
- Secondary alcohol
- Lactam
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Alcohol
- Organonitrogen compound
- Organic nitrogen compound
- Organooxygen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Primary alcohol
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05fu-9260000000-d61a359a55520b6ac4e6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0ayi-8888900000-435694604ab924019571 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0940000000-590c892f3434aeade64c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-5700e521158c2e2eebec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udr-1900000000-1f7aae4adebc1e5e4609 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001j-0790000000-63ac267157492bd6dacf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0900000000-d38b2649cc0a087967a3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-1900000000-54a59a321d15984ea2d3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-d3116843b691b79a17a6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-4c5f22a621f1bf5edca3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-1900000000-08bd7a023e7c71d7fa14 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000t-0950000000-e29272a7e2cfc8cd35b0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0900000000-5eddb3d0c408b01a8037 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-0900000000-7f4e88398d5f4a1895ce | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, CD3OD, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, Methanol, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CD3OD, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|