| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:03:29 UTC |
|---|
| Update Date | 2020-05-11 20:40:28 UTC |
|---|
| BMDB ID | BMDB0003290 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Gulonic acid |
|---|
| Description | Gulonic acid, also known as gulonate, belongs to the class of organic compounds known as medium-chain hydroxy acids and derivatives. These are hydroxy acids with a 6 to 12 carbon atoms long side chain. Based on a literature review a significant number of articles have been published on Gulonic acid. |
|---|
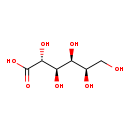
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Gulonate | Generator | | Gulonic acid, (L)-isomer | MeSH | | Gulonic acid, ion (1-), (L)-isomer | MeSH | | Gulonic acid, monosodium salt, (L)-isomer | MeSH | | D-Gulonate | HMDB | | D-Gulonic acid | HMDB | | L-Gulonate | HMDB | | L-Gulonic acid | HMDB |
|
|---|
| Chemical Formula | C6H12O7 |
|---|
| Average Molecular Weight | 196.1553 |
|---|
| Monoisotopic Molecular Weight | 196.058302738 |
|---|
| IUPAC Name | (2R,3R,4S,5R)-2,3,4,5,6-pentahydroxyhexanoic acid |
|---|
| Traditional Name | gulonic acid |
|---|
| CAS Registry Number | 20246-53-1 |
|---|
| SMILES | OC[C@@H](O)[C@H](O)[C@@H](O)[C@@H](O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H12O7/c7-1-2(8)3(9)4(10)5(11)6(12)13/h2-5,7-11H,1H2,(H,12,13)/t2-,3+,4-,5-/m1/s1 |
|---|
| InChI Key | RGHNJXZEOKUKBD-KKQCNMDGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as medium-chain hydroxy acids and derivatives. These are hydroxy acids with a 6 to 12 carbon atoms long side chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Hydroxy acids and derivatives |
|---|
| Sub Class | Medium-chain hydroxy acids and derivatives |
|---|
| Direct Parent | Medium-chain hydroxy acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Medium-chain hydroxy acid
- Medium-chain fatty acid
- Beta-hydroxy acid
- Hydroxy fatty acid
- Alpha-hydroxy acid
- Monosaccharide
- Fatty acyl
- Fatty acid
- Secondary alcohol
- Polyol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Primary alcohol
- Alcohol
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (6 TMS) | splash10-067l-0897000000-a5adc23ee8062f1b4496 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-067l-0897000000-a5adc23ee8062f1b4496 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-06tu-9500000000-abc0b019db16afd3352b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (6 TMS) - 70eV, Positive | splash10-03fr-6121297000-698854fa2453487a1d69 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_6_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Gulonic acid,6TBDMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0002-0900000000-51cc4bff4f073ec29dc7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-06yk-9200000000-36fd28057c23b3aeb698 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-01t9-9200000000-14a6bd042df22e1334ec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004j-2900000000-9081b4da0decbd05f2d0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-9400000000-79c4dcd310d5cacca35c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08i9-9000000000-62e401fc518adf3537bd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0adu-9600000000-2d2f2523012a7bbe0e8a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4u-9300000000-00e22f3157480a68e695 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-87d75121bafb2a63a0f8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9100000000-ff1473e44b436e4f8fa5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a6r-9000000000-aac8778b161b37e56a13 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6r-9000000000-8920759ff7b62f308018 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-024m-8900000000-7ecf18d54cb462c04558 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-074i-9000000000-a24dd2cb9b57deddeaae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08fr-9000000000-ae48df53c8de11d2cae6 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|