| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:06:19 UTC |
|---|
| Update Date | 2020-05-21 16:27:09 UTC |
|---|
| BMDB ID | BMDB0003848 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | D-Myo-inositol 3,4,5,6-tetrakisphosphate |
|---|
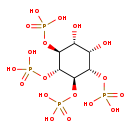
| Description | D-Myo-d-myo-inositol 3,4,5,6-tetrakisphosphate, also known as ins-3,4,5,6-P4 or d-myo-inositol 3,4,5,6-tetrakisphosphate, belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. D-Myo-d-myo-inositol 3,4,5,6-tetrakisphosphate is possibly soluble (in water) and an extremely strong acidic compound (based on its pKa). D-Myo-d-myo-inositol 3,4,5,6-tetrakisphosphate participates in a number of enzymatic reactions, within cattle. In particular, D-Myo-d-myo-inositol 3,4,5,6-tetrakisphosphate can be biosynthesized from inositol 1,3,4,5,6-pentakisphosphate; which is mediated by the enzyme inositol-tetrakisphosphate 1-kinase. In addition, D-Myo-d-myo-inositol 3,4,5,6-tetrakisphosphate can be biosynthesized from inositol 1,3,4,5,6-pentakisphosphate through the action of the enzyme inositol-tetrakisphosphate 1-kinase. In cattle, D-myo-d-myo-inositol 3,4,5,6-tetrakisphosphate is involved in a couple of metabolic pathways, which include the inositol metabolism pathway and the inositol phosphate metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Inositol-3,4,5,6-tetraphosphate | ChEBI | | Ins-3,4,5,6-P4 | ChEBI | | Myo-inositol-3,4,5,6-tetrakisphosphate | ChEBI | | Inositol 3,4,5,6-tetrakisphosphate | Kegg | | Inositol-3,4,5,6-tetraphosphoric acid | Generator | | Myo-inositol-3,4,5,6-tetrakisphosphoric acid | Generator | | Inositol 3,4,5,6-tetrakisphosphoric acid | Generator | | D-Myo-inositol 3,4,5,6-tetrakisphosphoric acid | Generator | | 1D-myo-Inositol 3,4,5,6-tetrakisphosphate | HMDB | | D-myo-Inositol, 3,4,5,6-tetrakis(dihydrogen phosphate) | HMDB | | Ins(3,4,5,6)P3 | MeSH, HMDB | | D-myo-Inositol 3,4,5,6-tetrakisphosphate | HMDB | | D-myo-Inositol 3,4,5,6-tetraphosphate | HMDB | | Inositol 3,4,5,6-tetrakis(phosphate) | HMDB | | Inositol 3,4,5,6-tetraphosphate | HMDB | | Ins(3,4,5,6)P4 | HMDB |
|
|---|
| Chemical Formula | C6H16O18P4 |
|---|
| Average Molecular Weight | 500.0755 |
|---|
| Monoisotopic Molecular Weight | 499.928709756 |
|---|
| IUPAC Name | {[(1R,2S,3R,4S,5S,6R)-3,4-dihydroxy-2,5,6-tris(phosphonooxy)cyclohexyl]oxy}phosphonic acid |
|---|
| Traditional Name | [(1R,2S,3R,4S,5S,6R)-3,4-dihydroxy-2,5,6-tris(phosphonooxy)cyclohexyl]oxyphosphonic acid |
|---|
| CAS Registry Number | 112791-61-4 |
|---|
| SMILES | O[C@H]1[C@@H](O)[C@H](OP(O)(O)=O)[C@@H](OP(O)(O)=O)[C@H](OP(O)(O)=O)[C@H]1OP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H16O18P4/c7-1-2(8)4(22-26(12,13)14)6(24-28(18,19)20)5(23-27(15,16)17)3(1)21-25(9,10)11/h1-8H,(H2,9,10,11)(H2,12,13,14)(H2,15,16,17)(H2,18,19,20)/t1-,2+,3-,4-,5+,6+/m0/s1 |
|---|
| InChI Key | MRVYFOANPDTYBY-UZAAGFTCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Inositol phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Inositol phosphate
- Monoalkyl phosphate
- Cyclohexanol
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Secondary alcohol
- 1,2-diol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | -7.268 | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9122600000-383a9e0d030e04f9efbc | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0fdk-9721484000-b7d5efdc25d786b39d17 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-5000790000-5390e100a7edacead23d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ue9-3000940000-553717cc294ea575f253 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pba-3109000000-2fceafe8a79fc5fd8156 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-3000900000-724d0d2ad6f25c90d193 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9002100000-815d445436dd2d0ccdbc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-2dc66a8603e5713437a3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0000090000-2a395f5fa8fd29e0fb49 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0000290000-2a13e4f6bebe4598fbd9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uk9-4309200000-6cea3cc017b0547b118b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000900000-43744c672ffce8e5f032 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000t-2000900000-1b7f098afeb3b6106811 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000100000-7111fc5de7c9d1b376ff | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Morgan Adam J; Komiya Shio; Xu Yingju; Miller Scott J Unified total syntheses of the inositol polyphosphates: D-I-3,5,6P3, D-I-3,4,5P3, D-I-3,4,6P3, and D-I-3,4,5,6P4 via catalytic enantioselective and site-selective phosphorylation. The Journal of organic chemistry (2006), 71(18), 6923-31. |
|---|