| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:08:34 UTC |
|---|
| Update Date | 2020-05-21 16:28:30 UTC |
|---|
| BMDB ID | BMDB0004220 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
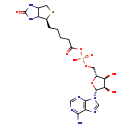
| Common Name | Biotinyl-5'-AMP |
|---|
| Description | Biotinyl-5'-AMP, also known as b-AMP or bio-5-AMP, belongs to the class of organic compounds known as 5'-acylphosphoadenosines. These are ribonucleoside derivatives containing an adenoside moiety, where the phosphate group is acylated. Biotinyl-5'-AMP is possibly soluble (in water) and a strong basic compound (based on its pKa). Biotinyl-5'-AMP exists in all living organisms, ranging from bacteria to humans. In cattle, biotinyl-5'-AMP is involved in the metabolic pathway called the biotin metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (+)-Biotinyl 5'-adenylate | HMDB | | 5'-Adenylic acid anhydride with biotin | HMDB | | 5'-Adenylic acid monoanhydride with biotin | HMDB | | b-AMP | HMDB | | beta-AMP | HMDB | | Bio-5-AMP | HMDB | | Biotin anhydride with 5'-adenylic acid | HMDB | | Biotinoyl 5'-adenylate | HMDB | | Biotinyl 5'-AMP | HMDB | | [3AS-(3aa,4b,6aa)]-5'-adenylic acid monoanhydride with hexahydro-2-oxo-1H-thieno[3,4-D]imidazole-4-pentanoate | HMDB | | [3AS-(3aa,4b,6aa)]-5'-adenylic acid monoanhydride with hexahydro-2-oxo-1H-thieno[3,4-D]imidazole-4-pentanoic acid | HMDB |
|
|---|
| Chemical Formula | C20H28N7O9PS |
|---|
| Average Molecular Weight | 573.517 |
|---|
| Monoisotopic Molecular Weight | 573.140682731 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}({5-[(4S)-2-oxo-hexahydro-1H-thieno[3,4-d]imidazolidin-4-yl]pentanoyl}oxy)phosphinic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy({5-[(4S)-2-oxo-hexahydrothieno[3,4-d]imidazolidin-4-yl]pentanoyl}oxy)phosphinic acid |
|---|
| CAS Registry Number | 4130-20-5 |
|---|
| SMILES | NC1=NC=NC2=C1N=CN2[C@@H]1O[C@H](COP(O)(=O)OC(=O)CCCC[C@@H]2SCC3NC(=O)NC23)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C20H28N7O9PS/c21-17-14-18(23-7-22-17)27(8-24-14)19-16(30)15(29)10(35-19)5-34-37(32,33)36-12(28)4-2-1-3-11-13-9(6-38-11)25-20(31)26-13/h7-11,13,15-16,19,29-30H,1-6H2,(H,32,33)(H2,21,22,23)(H2,25,26,31)/t9?,10-,11+,13?,15-,16-,19-/m1/s1 |
|---|
| InChI Key | UTQCSTJVMLODHM-CFYKWCKVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 5'-acylphosphoadenosines. These are ribonucleoside derivatives containing an adenoside moiety, where the phosphate group is acylated. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | 5'-acylphosphoadenosines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 5'-acylphosphoadenosine
- Biotin_derivative
- Biotin
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Thienoimidazolidine
- Acyl phosphate
- Aminopyrimidine
- Monoalkyl phosphate
- Imidazolidinone
- Imidolactam
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Pyrimidine
- Thiophene
- Thiolane
- Tetrahydrofuran
- Azole
- Imidazole
- Imidazolidine
- Heteroaromatic compound
- Amino acid or derivatives
- Carbonic acid derivative
- Urea
- Secondary alcohol
- 1,2-diol
- Carboxylic acid salt
- Dialkylthioether
- Oxacycle
- Azacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Thioether
- Organoheterocyclic compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organic oxygen compound
- Organic nitrogen compound
- Organic salt
- Organonitrogen compound
- Organooxygen compound
- Organopnictogen compound
- Organic oxide
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-6729030000-2cb82379b8e96b597b13 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00kb-9733703000-98bae5a17ded03704629 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Biotinyl-5'-AMP,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0942030000-e61e914c7ef0f87ef7ce | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0910000000-90a339c73ce7d6e18307 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2900000000-4473efe13a9fdcd356f6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00c0-2309030000-db991b74538e5511fc08 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-003r-7902000000-c38c4f3a31b23503bb6d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-003r-9400000000-f14ea50608a771c5ffc0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0101090000-26c429c0d542be98a853 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0940020000-3590ed25fbf2aee9ac0c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0931010000-189f936a74fee5be25d1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0001090000-4427dd4df762a2ebe889 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00b9-9004080000-1810ce9ed7993ec252eb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01t9-9200000000-0a1f3c98da5c89d7c119 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|