| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:10:47 UTC |
|---|
| Update Date | 2020-05-11 20:40:58 UTC |
|---|
| BMDB ID | BMDB0004824 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | N2,N2-Dimethylguanosine |
|---|
| Description | N2,N2-Dimethylguanosine, also known as M22G, belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. Based on a literature review a significant number of articles have been published on N2,N2-Dimethylguanosine. |
|---|
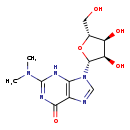
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2,2-Dimethylguanosine | ChEBI | | 2-(Dimethylamino)-9-(beta-D-ribofuranosyl)-1,9-dihydro-6H-purin-6-one | ChEBI | | 2-Dimethylamino-6-oxypurine riboside | ChEBI | | m22g | ChEBI | | N2-Dimethylguanosine | ChEBI | | 2-(Dimethylamino)-9-(b-D-ribofuranosyl)-1,9-dihydro-6H-purin-6-one | Generator | | 2-(Dimethylamino)-9-(β-D-ribofuranosyl)-1,9-dihydro-6H-purin-6-one | Generator | | N,N- Dimethylguanosine | HMDB | | N,N-Dimethyl-guanosine | HMDB | | m(2)2g | MeSH, HMDB | | m2(2)g | MeSH, HMDB | | N(2),N(2)-Dimethylguanosine | MeSH, HMDB | | m(2)(2)g | MeSH, HMDB | | N2,N2-Dimethylguanosine | MeSH |
|
|---|
| Chemical Formula | C12H17N5O5 |
|---|
| Average Molecular Weight | 311.2939 |
|---|
| Monoisotopic Molecular Weight | 311.122968679 |
|---|
| IUPAC Name | 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(dimethylamino)-6,9-dihydro-3H-purin-6-one |
|---|
| Traditional Name | 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(dimethylamino)-3H-purin-6-one |
|---|
| CAS Registry Number | 2140-67-2 |
|---|
| SMILES | CN(C)C1=NC(=O)C2=C(N1)N(C=N2)[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C12H17N5O5/c1-16(2)12-14-9-6(10(21)15-12)13-4-17(9)11-8(20)7(19)5(3-18)22-11/h4-5,7-8,11,18-20H,3H2,1-2H3,(H,14,15,21)/t5-,7-,8-,11-/m1/s1 |
|---|
| InChI Key | RSPURTUNRHNVGF-IOSLPCCCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleosides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Purine nucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleoside
- Glycosyl compound
- N-glycosyl compound
- Hypoxanthine
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Dialkylarylamine
- Aminopyrimidine
- Hydroxypyrimidine
- Monosaccharide
- N-substituted imidazole
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Tetrahydrofuran
- Secondary alcohol
- Azacycle
- Oxacycle
- Organoheterocyclic compound
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Hydrocarbon derivative
- Organopnictogen compound
- Organic oxygen compound
- Primary alcohol
- Alcohol
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 235 - 236 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ac3-9180000000-6b74485ca69b8a0937c1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-01c0-3391310000-26de80f9bc78e87a644a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-001i-1900000000-d79f213e790ad370a66e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0059-0900000000-20a29955f6b438cd3fa1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0901000000-1701dcf20f0234b63b6d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0309000000-1bbce8944068addfb261 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-004i-0900000000-95ebcc742789eafc2b3b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0309000000-d555efe7388f8c08d0e5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-004i-0900000000-4412ec0093a05d259e3c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0009000000-91ed5c716462a0de56ed | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - CE-QTOF-MS system (Agilent 7100 CE + 6550 QTOF) 10V, Positive | splash10-001i-0900000000-6033a973f510df066a11 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0901000000-ee7f601ba19e92eb3000 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-001i-0900000000-1ee81ff6672079088e87 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0904000000-49681e90d78457e6d0c9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-001i-0900000000-b320e8e065d43857d0a5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0900000000-8a37caef7033915dada7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-001i-0900000000-16a2c20e204fd9a3725c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 0V, Positive | splash10-03e9-0709000000-956e2532e241964d69e9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0900000000-0138cd5cbbbc494c7b3f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-f21ba6a3e1d687e0fe98 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-001i-3900000000-b511ad9d8a15f64f7f66 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-001i-0900000000-2391230a8b33e68d459b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 0V, Positive | splash10-03e9-0709000000-057ab6ac2589b80194f0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-ce6965fbbcad719a81b7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-001i-1900000000-fd4a6954268248ad2cdc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03fr-0759000000-2b4a326aad3ce87ea5e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0910000000-10561e691bbf8dac63d5 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|