| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:18:06 UTC |
|---|
| Update Date | 2020-03-13 16:39:45 UTC |
|---|
| BMDB ID | BMDB0005763 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Thyrotropin-releasing factor |
|---|
| Description | Thyrotropin releasing hormone, also known as TRH or protirelin, belongs to the class of organic compounds known as estrogens and derivatives. These are steroids with a structure containing a 3-hydroxylated estrane. Thyrotropin releasing hormone is a very strong basic compound (based on its pKa). |
|---|
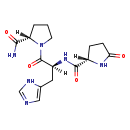
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| L-Pyroglutamyl-L-histidyl-L-prolineamide | ChEBI | | Thyroliberin | ChEBI | | Thyrotropic releasing hormone | ChEBI | | Thyrotropic-releasing factor | ChEBI | | Thyrotropin-releasing factor | ChEBI | | TRH | ChEBI | | TSH-Releasing factor | ChEBI | | TSH-Releasing hormone | ChEBI | | Protirelin | Kegg | | Relefact TRH | Kegg | | Abbott brand OF protirelin | HMDB | | Abbott-38579 | HMDB | | Antepan | HMDB | | Aventis brand OF protirelin | HMDB | | Novartis brand OF protirelin | HMDB | | Proterelin tartrate | HMDB | | Proterelin tartrate hydrate | HMDB | | Protirelin abbott brand | HMDB | | Protirelin aventis brand | HMDB | | Stimu TSH | HMDB | | Tartrate hydrate, proterelin | HMDB | | Thypinone | HMDB | | Abbott 38579 | HMDB | | Protirelin tartrate (1:1) | HMDB | | TRH Ferring | HMDB | | TRH Prem | HMDB | | Thyrotropin-releasing hormone | HMDB | | Thyrotropin-releasing hormone tartrate | HMDB | | Abbott38579 | HMDB | | Ferring brand OF protirelin | HMDB | | Henning berlin brand OF protirelin | HMDB | | Hydrate, proterelin tartrate | HMDB | | Merck brand OF protirelin | HMDB | | Prem, TRH | HMDB | | Protirelin ferring brand | HMDB | | Protirelin merck brand | HMDB | | Stimu-TSH | HMDB | | Thyroliberin TRH merck | HMDB | | Thyrotropin releasing factor | HMDB | | Protirelin novartis brand | HMDB | | StimuTSH | HMDB | | TRH, Relefact | HMDB | | Thyrotropin releasing hormone tartrate | HMDB | | Thyrotropin releasing hormone | ChEBI |
|
|---|
| Chemical Formula | C16H22N6O4 |
|---|
| Average Molecular Weight | 362.3837 |
|---|
| Monoisotopic Molecular Weight | 362.170253222 |
|---|
| IUPAC Name | (2S)-N-[(2S)-1-[(2S)-2-carbamoylpyrrolidin-1-yl]-3-(1H-imidazol-5-yl)-1-oxopropan-2-yl]-5-oxopyrrolidine-2-carboxamide |
|---|
| Traditional Name | (2S)-N-[(2S)-1-[(2S)-2-carbamoylpyrrolidin-1-yl]-3-(3H-imidazol-4-yl)-1-oxopropan-2-yl]-5-oxopyrrolidine-2-carboxamide |
|---|
| CAS Registry Number | 24305-27-9 |
|---|
| SMILES | [H][C@@](CC1=CN=CN1)(NC(=O)[C@]1([H])CCC(=O)N1)C(=O)N1CCC[C@@]1([H])C(N)=O |
|---|
| InChI Identifier | InChI=1S/C16H22N6O4/c17-14(24)12-2-1-5-22(12)16(26)11(6-9-7-18-8-19-9)21-15(25)10-3-4-13(23)20-10/h7-8,10-12H,1-6H2,(H2,17,24)(H,18,19)(H,20,23)(H,21,25)/t10-,11-,12-/m0/s1 |
|---|
| InChI Key | XNSAINXGIQZQOO-SRVKXCTJSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as estrogens and derivatives. These are steroids with a structure containing a 3-hydroxylated estrane. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Estrane steroids |
|---|
| Direct Parent | Estrogens and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Estrogen-skeleton
- 3-oxosteroid
- Hydroxysteroid
- Oxosteroid
- 17-hydroxysteroid
- 2-oxosteroid
- Cyclic alcohol
- Cyclic ketone
- Ketone
- Secondary alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-001i-9132000000-49fa8da4e6762d408997 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0ika-0196000000-afe7149844b51b017ffe | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0uxr-2890000000-b54b008a540dfef60574 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-06e9-4900000000-ee9272d8925098115bfd | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0uxr-1980000000-71c7b1b1167912a394e4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0229000000-07452232680491a34370 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03k9-4910000000-36df0cdebfe4ff6da0c5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-00xs-0590000000-a869c0d113c010b718fa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03di-4900000000-4508511e6a002d601f08 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-00xs-1690000000-48f23b21f382fe83e1ec | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03k9-7900000000-c0fe7f08cbc139f31fc5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-02mj-1794000000-14f46bfee6df7d4a5a00 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0229-3960000000-af2ed02667f34a7fcac6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0139000000-3e36ea4f46eaffc55495 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-00xs-0790000000-099b2f8d353b67c2e883 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ika-2369000000-319ede954f3ced479d6e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9582000000-bdc5d7fba5104a9bcfe8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00lr-9400000000-32c92be7b84a0dc3663b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-1129000000-e58ba2209f3459479f64 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-6968000000-2fff001bc2b3c86f57d8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9411000000-a08e549b33f1b6a81063 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0009000000-696431d5c8a5cf963eca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-044i-4439000000-6797de0104dd10461c61 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dj-9812000000-16c08d58395f971c0c3e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0019000000-f097eb2b9e44187f9db3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9762000000-3dbee1b2272386f8656e | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Matsuda, Fuyuhiko; Itoh, Shin; Hattori, Noritaka; Yanagiya, Mitsutoshi; Matsumoto, Takeshi. A simple method for synthesis of amides and peptides through acyl chlorides. A rapid synthesis of thyrotropin releasing hormone. Tetrahedron (1985), 41(18), 3625-3 |
|---|