| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:19:52 UTC |
|---|
| Update Date | 2020-04-22 15:16:45 UTC |
|---|
| BMDB ID | BMDB0006038 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
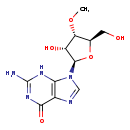
| Common Name | 3'-O-Methylguanosine |
|---|
| Description | 3'-O-Methylguanosine, also known as 3-OMG, belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. Based on a literature review a significant number of articles have been published on 3'-O-Methylguanosine. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-OMG | ChEBI | | 3'-O-Methyl-guanosine | HMDB |
|
|---|
| Chemical Formula | C11H15N5O5 |
|---|
| Average Molecular Weight | 297.2673 |
|---|
| Monoisotopic Molecular Weight | 297.107318615 |
|---|
| IUPAC Name | 2-amino-9-[(2R,3R,4S,5R)-3-hydroxy-5-(hydroxymethyl)-4-methoxyoxolan-2-yl]-6,9-dihydro-3H-purin-6-one |
|---|
| Traditional Name | 2-amino-9-[(2R,3R,4S,5R)-3-hydroxy-5-(hydroxymethyl)-4-methoxyoxolan-2-yl]-3H-purin-6-one |
|---|
| CAS Registry Number | 10300-27-3 |
|---|
| SMILES | CO[C@@H]1[C@@H](CO)O[C@H]([C@@H]1O)N1C=NC2=C1NC(N)=NC2=O |
|---|
| InChI Identifier | InChI=1S/C11H15N5O5/c1-20-7-4(2-17)21-10(6(7)18)16-3-13-5-8(16)14-11(12)15-9(5)19/h3-4,6-7,10,17-18H,2H2,1H3,(H3,12,14,15,19)/t4-,6-,7-,10-/m1/s1 |
|---|
| InChI Key | UYARPHAXAJAZLU-KQYNXXCUSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleosides. Purine nucleosides are compounds comprising a purine base attached to a ribosyl or deoxyribosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleosides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Purine nucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleoside
- Glycosyl compound
- N-glycosyl compound
- Imidazopyrimidine
- Purine
- Hydroxypyrimidine
- Monosaccharide
- N-substituted imidazole
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Tetrahydrofuran
- Secondary alcohol
- Oxacycle
- Ether
- Azacycle
- Organoheterocyclic compound
- Dialkyl ether
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Hydrocarbon derivative
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 263 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-007c-9320000000-a45c28e7d4a162bff5e6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0592-4921000000-ca1307a112db163864a5 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-03dj-0910000000-2a344b5dc7761621cdb0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0940000000-18f0214f63b3ea1712a2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-ee3c0cf719ddeda0ce90 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udr-0900000000-165eaae08a8f4384a473 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0f6t-0490000000-abe3f6b4224f76721a77 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0900000000-5d454c5b699b2630b62b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0pbc-4900000000-b26752858470579e4fe8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-361a2474138028086bb3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-e4f63a292fa80bd152db | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f79-1900000000-df57ad6bf2c6c29c8535 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0290000000-bf93be1dd1c5bd53bc36 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0920000000-e776feba748389e952d6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0kal-3900000000-ade1ae25004ec1352c2d | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|