| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:20:38 UTC |
|---|
| Update Date | 2020-05-21 16:29:05 UTC |
|---|
| BMDB ID | BMDB0006229 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 5-Diphosphoinositol pentakisphosphate |
|---|
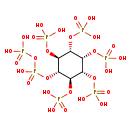
| Description | 5-Diphosphoinositol pentakisphosphate, also known as 5-diphosphoinositol pentakisphosphate, belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. 5-Diphosphoinositol pentakisphosphate is possibly soluble (in water) and an extremely strong acidic compound (based on its pKa). 5-Diphosphoinositol pentakisphosphate exists in all eukaryotes, ranging from yeast to humans. 5-Diphosphoinositol pentakisphosphate participates in a number of enzymatic reactions, within cattle. In particular, 5-Diphosphoinositol pentakisphosphate can be biosynthesized from bisdiphosphoinositol tetrakisphosphate through its interaction with the enzyme diphosphoinositol polyphosphate phosphohydrolase 1. In addition, 5-Diphosphoinositol pentakisphosphate can be converted into bisdiphosphoinositol tetrakisphosphate; which is mediated by the enzyme inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 1. In cattle, 5-diphosphoinositol pentakisphosphate is involved in the metabolic pathway called the inositol phosphate metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (1R,2R,3S,4S,5R,6S)-2,3,4,5,6-Pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate | ChEBI | | 1D-Myo-inositol 5-diphosphate 1,2,3,4,6-pentakisphosphate | ChEBI | | 1D-Myo-inositol 5-diphosphate pentakisphosphate | ChEBI | | 5-PP-InsP5 | ChEBI | | (1R,2R,3S,4S,5R,6S)-2,3,4,5,6-Pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphoric acid | Generator | | 1D-Myo-inositol 5-diphosphoric acid 1,2,3,4,6-pentakisphosphoric acid | Generator | | 1D-Myo-inositol 5-diphosphoric acid pentakisphosphoric acid | Generator | | 5-Diphosphoinositol pentakisphosphoric acid | Generator | | 1,2,3,4,6-Pentakis-O-phosphono-1D-myo-inositol 5-(trihydrogen diphosphate) | HMDB | | 5-Diphospho-1D-myo-inositol 1,2,3,4,6-pentakisphosphate | HMDB | | 5-Diphospho-1D-myo-inositol pentakisphosphate | HMDB | | 5beta 5pp-IP5 | HMDB | | 5beta-IP7 | HMDB | | Diphosphoinositol pentakisphosphate | HMDB | | InsP7 | HMDB | | IP7 | HMDB | | PP-InsP5 | HMDB | | myo-Inositol 1,2,3,4,6-pentakis(dihydrogen phosphate) 5-(trihydrogen diphosphate) | HMDB |
|
|---|
| Chemical Formula | C6H19O27P7 |
|---|
| Average Molecular Weight | 740.0152 |
|---|
| Monoisotopic Molecular Weight | 739.827700986 |
|---|
| IUPAC Name | {[(1R,2r,3S,4R,5s,6S)-2-{[hydroxy(phosphonooxy)phosphoryl]oxy}-3,4,5,6-tetrakis(phosphonooxy)cyclohexyl]oxy}phosphonic acid |
|---|
| Traditional Name | [(1R,2r,3S,4R,5s,6S)-2-{[hydroxy(phosphonooxy)phosphoryl]oxy}-3,4,5,6-tetrakis(phosphonooxy)cyclohexyl]oxyphosphonic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | OP(O)(=O)O[C@@H]1[C@H](OP(O)(O)=O)[C@@H](OP(O)(O)=O)[C@H](OP(O)(=O)OP(O)(O)=O)[C@@H](OP(O)(O)=O)[C@@H]1OP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H19O27P7/c7-34(8,9)27-1-2(28-35(10,11)12)4(30-37(16,17)18)6(32-40(25,26)33-39(22,23)24)5(31-38(19,20)21)3(1)29-36(13,14)15/h1-6H,(H,25,26)(H2,7,8,9)(H2,10,11,12)(H2,13,14,15)(H2,16,17,18)(H2,19,20,21)(H2,22,23,24)/t1-,2+,3-,4-,5+,6+ |
|---|
| InChI Key | UPHPWXPNZIOZJL-KXXVROSKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Inositol phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Inositol phosphate
- Organic pyrophosphate
- Monoalkyl phosphate
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-056v-3500129000-92e02f48f80539fd2ebb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ox-2200039500-ca3324ec34611cb741e8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01ox-1100019100-a4bf41f87596bb42e4aa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-3200592000-718743d53ee9b1a54936 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002r-4100001900-58fc5d06ef574febc5fc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-9300013300-99dcc5e94c530a9c5edd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-7891e87bda74c3a75d76 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0000000900-4db44a159198f14597ed | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00bi-5000001900-ae421193acbffe665198 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9100013000-12bcf1f710fd737c956b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000000900-93209a4c8cd3c7e5e96b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-0000009500-e5f85269addd70fd0c57 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01q9-0000950000-7df113ce88ed8658d548 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|