| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:35:45 UTC |
|---|
| Update Date | 2020-04-22 15:49:32 UTC |
|---|
| BMDB ID | BMDB0012303 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | UDP-L-arabinose |
|---|
| Description | UDP-L-arabinose, also known as UDP-beta-L-arap or UDP-b-L-arabinose, belongs to the class of organic compounds known as pyrimidine ribonucleoside diphosphates. These are pyrimidine ribonucleotides with diphosphate group linked to the ribose moiety. Based on a literature review a significant number of articles have been published on UDP-L-arabinose. |
|---|
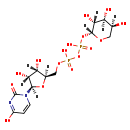
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| UDP-beta-L-Arabinose | ChEBI | | UDP-beta-L-Arap | ChEBI | | Uridine 5'-diphospho-beta-L-arabinopyranose | ChEBI | | UDP-b-L-Arabinose | Generator | | UDP-Β-L-arabinose | Generator | | UDP-b-L-Arap | Generator | | UDP-Β-L-arap | Generator | | Uridine 5'-diphospho-b-L-arabinopyranose | Generator | | Uridine 5'-diphospho-β-L-arabinopyranose | Generator | | UDP-b-L-Arabinopyranose | Generator | | UDP-β-L-arabinopyranose | Generator | | UDP-L-arabinose | HMDB | | UDP-arabinose | HMDB | | UDP-beta-L-arabinopyranose | HMDB | | Uridine 5'-(trihydrogen diphosphate) mono-L-arabinopyranosyl ester | HMDB | | Uridine 5'-(trihydrogen pyrophosphate) mono-L-arabinopyranosyl ester | HMDB | | Uridine 5'-(trihydrogen pyrophosphate) mono-L-arabinosyl ester | HMDB | | Uridine 5'-diphosphate arabinoside | HMDB | | Uridine 5'-pyrophosphate, beta-L-arabinosyl ester | HMDB | | Uridine 5'-pyrophosphate, β-L-arabinosyl ester | HMDB | | Uridine 5’-(trihydrogen diphosphate) mono-L-arabinopyranosyl ester | HMDB | | Uridine 5’-(trihydrogen pyrophosphate) mono-L-arabinopyranosyl ester | HMDB | | Uridine 5’-(trihydrogen pyrophosphate) mono-L-arabinosyl ester | HMDB | | Uridine 5’-diphosphate arabinoside | HMDB | | Uridine 5’-pyrophosphate, β-L-arabinosyl ester | HMDB | | Uridine diphosphate arabinose | HMDB | | Uridine diphospho-beta-L-arabinopyranose | HMDB | | Uridine diphospho-β-L-arabinopyranose | HMDB | | Uridine diphosphoarabinose | HMDB |
|
|---|
| Chemical Formula | C14H22N2O16P2 |
|---|
| Average Molecular Weight | 536.2758 |
|---|
| Monoisotopic Molecular Weight | 536.04445569 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-hydroxy-2-oxo-1,2-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy}({[hydroxy({[(2R,3R,4S,5S)-3,4,5-trihydroxyoxan-2-yl]oxy})phosphoryl]oxy})phosphinic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-hydroxy-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy([hydroxy([(2R,3R,4S,5S)-3,4,5-trihydroxyoxan-2-yl]oxy)phosphoryl]oxy)phosphinic acid |
|---|
| CAS Registry Number | 15839-78-8 |
|---|
| SMILES | O[C@@H]1[C@@H](COP(O)(=O)OP(O)(=O)O[C@H]2OC[C@H](O)[C@H](O)[C@H]2O)O[C@H]([C@@H]1O)N1C=CC(=O)NC1=O |
|---|
| InChI Identifier | InChI=1S/C14H22N2O16P2/c17-5-3-28-13(11(22)8(5)19)31-34(26,27)32-33(24,25)29-4-6-9(20)10(21)12(30-6)16-2-1-7(18)15-14(16)23/h1-2,5-6,8-13,17,19-22H,3-4H2,(H,24,25)(H,26,27)(H,15,18,23)/t5-,6+,8-,9+,10+,11+,12+,13+/m0/s1 |
|---|
| InChI Key | DQQDLYVHOTZLOR-IAZOVDBXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine ribonucleoside diphosphates. These are pyrimidine ribonucleotides with diphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine ribonucleotides |
|---|
| Direct Parent | Pyrimidine ribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine ribonucleoside diphosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Organic pyrophosphate
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Monosaccharide
- Organic phosphoric acid derivative
- Oxane
- Phosphoric acid ester
- Pyrimidine
- Alkyl phosphate
- Vinylogous amide
- Heteroaromatic compound
- Tetrahydrofuran
- Lactam
- Urea
- Secondary alcohol
- Oxacycle
- Polyol
- Organoheterocyclic compound
- Azacycle
- Alcohol
- Organic nitrogen compound
- Organic oxide
- Organonitrogen compound
- Organooxygen compound
- Organic oxygen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-016r-4936760000-00f37a38fe624376e23d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0300-4642229000-26e6ed2c6eb4497585c2 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0901110000-8ae0db777cff3d445c4c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-4910000000-67006286cf564f62ff42 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-5900000000-da262c67a1973399a755 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000f-8713790000-946e0081f7ad1a09d3d6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ox-9622010000-7089747eea74980128f4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06tf-4900000000-53eebd3beb66a7fd8d3f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0001090000-5cb9330b784831a318c0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0btc-5913210000-b3ea7476b5c2e48ecb1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-4956200000-d7160be3d1a922677c83 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dr-1800690000-ee1b02fe5fb340e69a4b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-4961210000-998fdbdb53c26f8dfaa2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03ea-5921000000-c5798a971977016536d7 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|