| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-03 19:35:40 UTC |
|---|
| Update Date | 2020-04-22 15:56:32 UTC |
|---|
| BMDB ID | BMDB0064012 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Methionyl-Tryptophan |
|---|
| Description | Methionyl-Tryptophan, also known as m-W dipeptide or met-TRP, belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. Based on a literature review very few articles have been published on Methionyl-Tryptophan. |
|---|
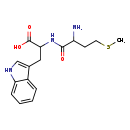
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| L-Methionyl-L-tryptophan | HMDB | | m-W Dipeptide | HMDB | | Met-TRP | HMDB | | Methionine tryptophan dipeptide | HMDB | | Methionine-tryptophan dipeptide | HMDB | | Methionyltryptophan | HMDB | | MW Dipeptide | HMDB | | 2-{[2-amino-1-hydroxy-4-(methylsulfanyl)butylidene]amino}-3-(1H-indol-3-yl)propanoate | HMDB | | 2-{[2-amino-1-hydroxy-4-(methylsulphanyl)butylidene]amino}-3-(1H-indol-3-yl)propanoate | HMDB | | 2-{[2-amino-1-hydroxy-4-(methylsulphanyl)butylidene]amino}-3-(1H-indol-3-yl)propanoic acid | HMDB | | Methionyl-tryptophan | MeSH |
|
|---|
| Chemical Formula | C16H21N3O3S |
|---|
| Average Molecular Weight | 335.421 |
|---|
| Monoisotopic Molecular Weight | 335.130362243 |
|---|
| IUPAC Name | 2-[2-amino-4-(methylsulfanyl)butanamido]-3-(1H-indol-3-yl)propanoic acid |
|---|
| Traditional Name | met-trp |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CSCCC(N)C(=O)NC(CC1=CNC2=CC=CC=C12)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C16H21N3O3S/c1-23-7-6-12(17)15(20)19-14(16(21)22)8-10-9-18-13-5-3-2-4-11(10)13/h2-5,9,12,14,18H,6-8,17H2,1H3,(H,19,20)(H,21,22) |
|---|
| InChI Key | XYVRXLDSCKEYES-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- Methionine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Alpha-amino acid amide
- Indolyl carboxylic acid derivative
- Alpha-amino acid or derivatives
- 3-alkylindole
- Indole
- Indole or derivatives
- Fatty amide
- N-acyl-amine
- Substituted pyrrole
- Benzenoid
- Fatty acyl
- Pyrrole
- Heteroaromatic compound
- Amino acid or derivatives
- Carboxamide group
- Amino acid
- Secondary carboxylic acid amide
- Organoheterocyclic compound
- Dialkylthioether
- Sulfenyl compound
- Azacycle
- Thioether
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Primary aliphatic amine
- Amine
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Carbonyl group
- Organic oxygen compound
- Hydrocarbon derivative
- Primary amine
- Organonitrogen compound
- Organooxygen compound
- Organosulfur compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0f8i-9840000000-316198d98347e3c4926f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0nu3-9424000000-3f883f4cb9d2d06b12f7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fs9-0839000000-977e1bd91be32ac7d1b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0pbi-4921000000-1a93bd08572f04ede59d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0kar-9600000000-75a4dc8a77c048aa28ed | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001j-7059000000-3e7d9a75f0df916abbe5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9121000000-0c8888f5d842b5fba781 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-9300000000-65e45321163e370369cf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0009000000-96ebb34e72cf67967f1d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-007k-9543000000-d4c4185cb07682b5ef8e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014j-4910000000-e777bf20ec0784a67c20 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0139000000-85c26c4b26fb23efa19a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052u-2693000000-9555d1fb08996cc1d7ed | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0aor-9600000000-da30b6573d8f369985d3 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|