| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-26 13:17:52 UTC |
|---|
| Update Date | 2020-04-22 20:21:34 UTC |
|---|
| BMDB ID | BMDB0109605 |
|---|
| Secondary Accession Numbers | None |
|---|
| Metabolite Identification |
|---|
| Common Name | Nonanoyl-CoA |
|---|
| Description | Nonanoyl-CoA belongs to the class of organic compounds known as very long-chain fatty acids. These are fatty acids with an aliphatic tail that contains at least 22 carbon atoms. Based on a literature review very few articles have been published on Nonanoyl-CoA. |
|---|
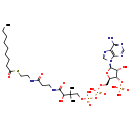
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| coenzyme A S-Nonanoate | HMDB | | coenzyme A S-Nonanoic acid | HMDB | | Nonanoyl-coenzyme A | HMDB | | 4-({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-2-hydroxy-3,3-dimethyl-N-(2-{[2-(nonanoylsulfanyl)ethyl]-C-hydroxycarbonimidoyl}ethyl)butanimidate | Generator | | 4-({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-2-hydroxy-3,3-dimethyl-N-(2-{[2-(nonanoylsulphanyl)ethyl]-C-hydroxycarbonimidoyl}ethyl)butanimidate | Generator | | 4-({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-2-hydroxy-3,3-dimethyl-N-(2-{[2-(nonanoylsulphanyl)ethyl]-C-hydroxycarbonimidoyl}ethyl)butanimidic acid | Generator |
|
|---|
| Chemical Formula | C30H52N7O17P3S |
|---|
| Average Molecular Weight | 907.757 |
|---|
| Monoisotopic Molecular Weight | 907.235323499 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({[hydroxy({3-hydroxy-2,2-dimethyl-3-[(2-{[2-(nonanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]propoxy})phosphoryl]oxy})phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-({[hydroxy([hydroxy(3-hydroxy-2,2-dimethyl-3-[(2-{[2-(nonanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]propoxy)phosphoryl]oxy)phosphoryl]oxy}methyl)oxolan-3-yl]oxyphosphonic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CCCCCCCCC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C30H52N7O17P3S/c1-4-5-6-7-8-9-10-21(39)58-14-13-32-20(38)11-12-33-28(42)25(41)30(2,3)16-51-57(48,49)54-56(46,47)50-15-19-24(53-55(43,44)45)23(40)29(52-19)37-18-36-22-26(31)34-17-35-27(22)37/h17-19,23-25,29,40-41H,4-16H2,1-3H3,(H,32,38)(H,33,42)(H,46,47)(H,48,49)(H2,31,34,35)(H2,43,44,45)/t19-,23-,24-,25?,29-/m1/s1 |
|---|
| InChI Key | WLDUTYVSAGSKIV-YUXRTGJASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as very long-chain fatty acids. These are fatty acids with an aliphatic tail that contains at least 22 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Very long-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Very long-chain fatty acid
- Unsaturated fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cell membrane
- Cytoplasm
- Membrane
|
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1920100120-197e2cc7bf0ee15417cf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0940300000-cdf29ead595810f66dd9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1910100000-e0160082789d90a03817 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-055r-2911030312-5e09ae40f908b844ee36 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001r-3911010000-42b5de3585c7978a8317 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057i-5900100000-eda134609e85dca13ffd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000000009-73a36281420219efecf3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a70-4500303649-513f54bc6c9e734635f4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00n0-5404501941-6f2d0eb01e44bc5e2fcb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0000000019-0887987085ff820f588f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0910101572-dba8175bda78f9c4cfe0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-0111900000-8797fa5ef7c0b52faca0 | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|