| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:26:49 UTC |
|---|
| Update Date | 2020-05-21 16:28:57 UTC |
|---|
| BMDB ID | BMDB0000213 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Myo-inositol 1-phosphate |
|---|
| Description | 1D-Myo-inositol 1-phosphate, also known as inositol 3-phosphoric acid, belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. 1D-Myo-inositol 1-phosphate is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). 1D-Myo-inositol 1-phosphate exists in all living species, ranging from bacteria to humans. |
|---|
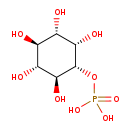
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1D-Myo-inositol 3-monophosphate | ChEBI | | 1l-Myo-inositol 1-phosphate | ChEBI | | D-Myo-inositol 3-monophosphate | ChEBI | | Inositol 3-phosphate | ChEBI | | L-Myo-inositol 1-phosphate | ChEBI | | Myoinositol 3-phosphate | ChEBI | | Myo-inositol 3-phosphate | Kegg | | Myo-inositol 3-monophosphate | Kegg | | Inositol 3-monophosphate | Kegg | | 1D-Myo-inositol 3-monophosphoric acid | Generator | | 1l-Myo-inositol 1-phosphoric acid | Generator | | D-Myo-inositol 3-monophosphoric acid | Generator | | Inositol 3-phosphoric acid | Generator | | L-Myo-inositol 1-phosphoric acid | Generator | | Myoinositol 3-phosphoric acid | Generator | | Myo-inositol 3-phosphoric acid | Generator | | Myo-inositol 3-monophosphoric acid | Generator | | Inositol 3-monophosphoric acid | Generator | | D-Myo-inositol 3-phosphoric acid | Generator | | Myo-inositol 1-phosphoric acid | HMDB | | 1-(Dihydrogen phosphate) DL-myo-inositol | HMDB | | 1-(Dihydrogen phosphate) myo-inositol | HMDB | | DL-Myo-inositol 1-phosphate | HMDB | | Myo-inositol 1-monophosphate | HMDB | | Myo-inositol-1-phosphate | HMDB | | Myoinositol 1-phosphate | HMDB | | 1D-Myo-inositol 1-phosphoric acid | HMDB | | D-myo-Inositol 3-phosphate | ChEBI |
|
|---|
| Chemical Formula | C6H13O9P |
|---|
| Average Molecular Weight | 260.1358 |
|---|
| Monoisotopic Molecular Weight | 260.029718526 |
|---|
| IUPAC Name | {[(1R,2S,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphonic acid |
|---|
| Traditional Name | myo-inositol 1-phosphate |
|---|
| CAS Registry Number | 573-35-3 |
|---|
| SMILES | O[C@H]1[C@H](O)[C@H](O)[C@H](OP(O)(O)=O)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H13O9P/c7-1-2(8)4(10)6(5(11)3(1)9)15-16(12,13)14/h1-11H,(H2,12,13,14)/t1-,2-,3+,4-,5-,6-/m0/s1 |
|---|
| InChI Key | INAPMGSXUVUWAF-PTQMNWPWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Inositol phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Inositol phosphate
- Monoalkyl phosphate
- Cyclohexanol
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Secondary alcohol
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 195 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 333.0 mg/mL | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (7 TMS) | splash10-014i-1559000000-8a2b9550f104f28e0f7d | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (7 TMS) | splash10-014i-0149000000-db7a2abef248fd1db09b | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-014i-1559000000-8a2b9550f104f28e0f7d | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-014i-0149000000-db7a2abef248fd1db09b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0005-9650000000-12db07845ff78aa5511d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-0a4i-2601259000-2775d0992be57f9b2782 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-2390000000-872b3b8044fff0ad80a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-2290000000-122539b694f8b31100a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9700000000-bed069cf85feb1dc4ab8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-4190000000-ea44c00eea91f419e692 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9220000000-d6c08f18832b9de73a53 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-3c92a73868c327380526 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0090000000-05d6010bb4bbf2b0c228 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a6s-9280000000-69bd06f4ab8866ad9db1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004j-9000000000-f3e6675946c65f8f8f07 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0090000000-89fa9427c518aa95bbd0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0190000000-fa4c1ecfe995d290a153 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9100000000-fc4055901243469636ad | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Klyashchitskii, B. A.; Pimenova, V. V.; Shvets, V. I.; Sokolov, S. D.; Preobrazhenskii, N. A. Total synthesis of sn-myoinositol 1-phosphate. Zhurnal Obshchei Khimii (1969), 39(10), 2373. |
|---|
| General References | - Melzer N, Wittenburg D, Hartwig S, Jakubowski S, Kesting U, Willmitzer L, Lisec J, Reinsch N, Repsilber D: Investigating associations between milk metabolite profiles and milk traits of Holstein cows. J Dairy Sci. 2013 Mar;96(3):1521-34. doi: 10.3168/jds.2012-5743. [PubMed:23438684 ]
|
|---|